ETL-Project
Home | BPMN Model | Use Case Model | ETL Pipeline | Insights | Team Contributions | About
ETL Pipeline Documentation
ETL Pipeline Documentation
1. Overview
The ETL (Extract, Transform, Load) pipeline is a structured process to automate the movement of healthcare data between systems. It ensures that patient and condition data is retrieved, cleaned, formatted, and loaded into a target system in a consistent and reliable manner. This pipeline focuses on API interactions and adheres to healthcare data standards like HL7 FHIR and SNOMED CT.
Pipeline Components
- Extraction: Retrieving patient and condition data from a source FHIR-compliant API.
- Transformation: Cleaning, structuring, and enriching the extracted data to meet the requirements of the target API.
- Loading: Posting the transformed data into the target API, while ensuring data integrity.
Extraction
- Uses Python to connect to FHIR APIs.
Extraction Process
The extraction process involved retrieving data from two main APIs:
1. FHIR Server for OpenEMR
- Base URL: https://in-info-web20.luddy.indianapolis.iu.edu/apis/default/fhir
- Endpoints:
/Patient: Retrieve patient details by ID or query parameters./Condition: Fetch patient-specific medical conditions.
- Authentication:
- Used OAuth 2.0 for authentication.
- Token management was performed by accessing the endpoint:
- Token URL: https://in-info-web20.luddy.indianapolis.iu.edu/oauth2/default/token
- Refresh tokens were stored and updated using utility scripts.
2. SNOMED CT
- Base URL: http://159.65.173.51:8080/v1/snomed
- Endpoints:
/concepts/{concept_id}: Retrieve basic information for a given SNOMED concept./concepts/{concept_id}/extended: Retrieves hierarchical parent relationships of the given SNOMED CT concept./concepts/{concept_id}/descriptions: Fetches the descriptions or terms associated with a specific SNOMED CT concept.
- Authentication: No authentication was required.
Transformation
- Data is cleaned and formatted for the target API.
Transformation Phase
The transformation phase ensures data is clean, standardized, and formatted to match the target API’s schema. This phase involves:
- Validation and Normalization: Ensuring fields like dates (ISO 8601) and addresses are consistent and error-free.
- Clinical Data Mapping: Mapping clinical data to SNOMED CT codes using ECL queries for standardization.
- Handling Missing/Invalid Values: Addressing incomplete or incorrect data to maintain integrity.
The following tools and techniques are integral to the transformation process:
- Requests Library: Used for interacting with APIs, sending data, and retrieving responses.
- Print Function: Extensively utilized for debugging and verifying data transformations.
- Python Libraries:
- json: Streamlines data parsing and formatting.
- datetime: Facilitates date manipulation and formatting.
Loading
- Processed data is posted back to the target API.
Loading Phase
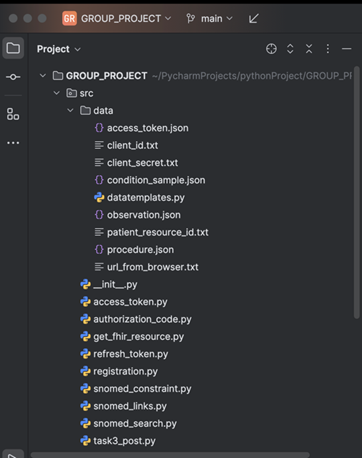
The src package in our project serves as a centralized location for core functionality, including data extraction, transformation, and loading (ETL). It ensures modularity, ease of maintenance, and clear organization of project-specific modules and scripts.
The following snippet illustrates the arrangement of our project for the Loading phase.
Code Demonstration: Patient Resource Creation
The below code demonstrates creating a patient resource in Primary Care EHR by using extracted data from FHIR. It saves the patient resource ID in patient_resource_id.txt for further processing and associates a condition with the created patient by transforming the SNOMED concept ID to its parent concept. The code also processes the second condition if available.
# Importing required modules
from data.datatemplates import patient_template_dict, condition_template_dict
from src.registration import data_dir
from src.snomed_links import constraint_parent, expression_constraint
# Base URLs
BASE_URL = "https://in-info-web20.luddy.indianapolis.iu.edu/apis/default/fhir"
BASE_PRIMARY_CARE_URL = "http://137.184.71.65:8080/fhir"
BASE_HERMES_URL = "http://159.65.173.51:8080/v1/snomed"
# Function to get access token from a file
def get_access_token_from_file():
...
# Function to get headers
def get_headers():
...
# Function to get patient resource ID
def get_patient_resource_id():
file_path = data_dir / "patient_resource_id.txt"
try:
with open(file_path, 'r') as file:
resource_id = file.read().strip()
return resource_id
except FileNotFoundError:
print(f"Error: {file_path} not found.")
return None
Task 1: Patient Template Dictionary
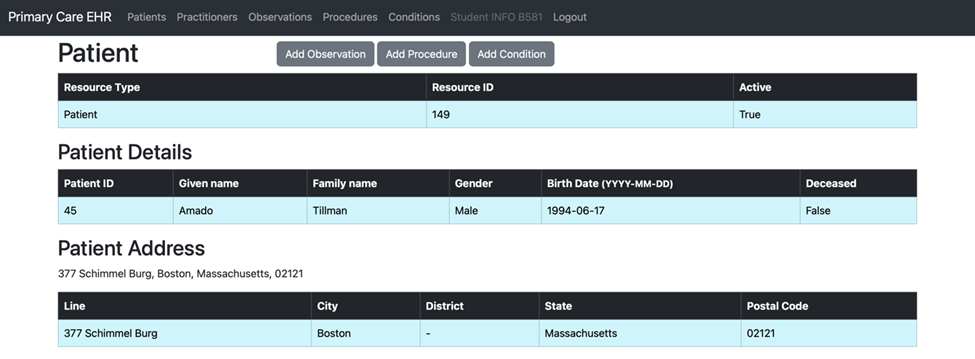
We created the patient_template_dict in the data directory to serve as a predefined structure for patient data. This template is essential for transforming raw data into the required format for submission to the Primary Care EHR, ensuring compliance with the FHIR standard.
def get_fhir_patient(resource_id):
url = f'{BASE_URL}/Patient/{resource_id}'
response = requests.get(url=url, headers=get_headers())
if response.status_code != 200:
print(f"Failed to fetch patient. Status Code: {response.status_code}")
print(f"Response Text: {response.text}")
return
data = response.json()
print("Response Data:", data) # Debugging: Inspect the response structure
# Check if 'name' field exists
name_list = data.get("name", [])
if not name_list:
print("Error: 'name' field is missing or empty in the response.")
return
birth_date = data.get('birthDate')
family_name = data['name'][0]['family']
given_name = data['name'][0]['given'][0]
possible_integers = [1, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50]
address = data.get('address', [{}])[0]
line = address.get('line', [''])[0]
city = address.get('city', '')
district = address.get('district', '-')
state = address.get('state', '')
postal_code = address.get('postalCode', '')
text = f"{line}, {city}, {state}, {postal_code}"
unique_patient_id = random.choice(possible_integers)
today_date = datetime.today().date().isoformat()
gender = data.get('gender')
# Populate the patient template
patient_template_dict["birthDate"] = birth_date
patient_template_dict["name"][0]["family"] = family_name
patient_template_dict["name"][0]["given"][0] = given_name
patient_template_dict["address"][0]["line"][0] = line
patient_template_dict["address"][0]["city"] = city
patient_template_dict["address"][0]["district"] = district
patient_template_dict["address"][0]["state"] = state
patient_template_dict["address"][0]["postalCode"] = postal_code
patient_template_dict["identifier"][0]["period"]["start"] = today_date
patient_template_dict["identifier"][0]["value"] = unique_patient_id
patient_template_dict["text"] = text
patient_template_dict["gender"] = gender
patient_template_dict["Address"][0]["text"] = text
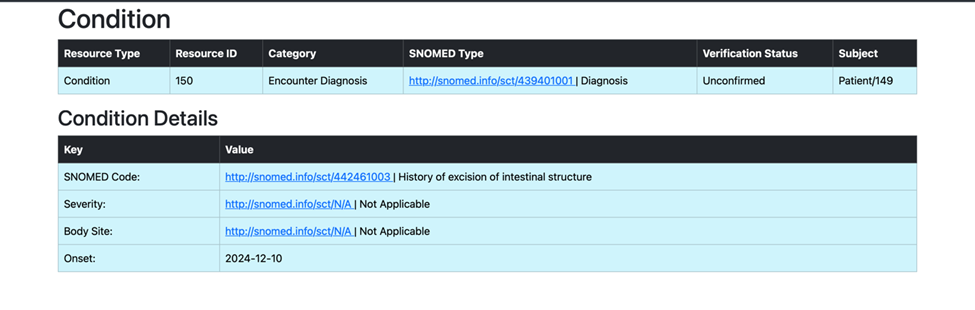
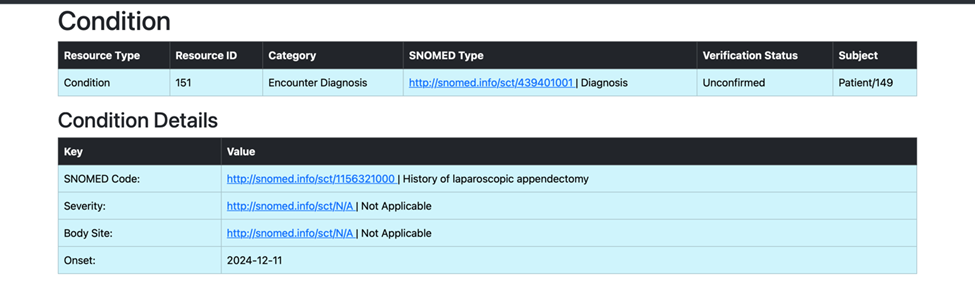
Task 2: Processing and Uploading Second Condition
With the use of the patient_resource_id.txt file, which stores the generated Patient Resource ID from the first task, we successfully retrieved and processed the second medical condition associated with the patient. Utilizing the SNOMED code from the second condition, we identified and created its corresponding child concept term. This enriched clinical detail was then transformed into a structured format using the predefined template and uploaded to the Primary Care EHR system as a new condition entry, ensuring accurate and comprehensive patient data representation.
def search_condition_second_child(patient_resource_id):
"""
Retrieve the second condition for a patient and post a new condition with the child concept term
"""
url = f'{BASE_URL}/Condition?patient={patient_resource_id}'
response = requests.get(url=url, headers=get_headers())
if response.status_code == 200:
data = response.json()
if 'entry' in data:
conditions = data['entry']
# Check if there are at least two conditions
if len(conditions) > 1:
second_condition = conditions[1] # Retrieve the second condition
snomed_code_from_openemr = second_condition["resource"]["code"]["coding"][0]["code"]
print(f"Retrieved SNOMED code from the second condition: {snomed_code_from_openemr}")
child_constraint = constraint_child(concept_id=snomed_code_from_openemr)
child_concept_id, child_concept_term = expression_constraint(search_string=child_constraint)
print(f"Identified Child Concept ID: {child_concept_id}")
print(f"Identified Child Preferred Term: {child_concept_term}")
condition_template_dict["code"]["text"] = child_concept_term
condition_template_dict["code"]["coding"][0]["display"] = child_concept_term
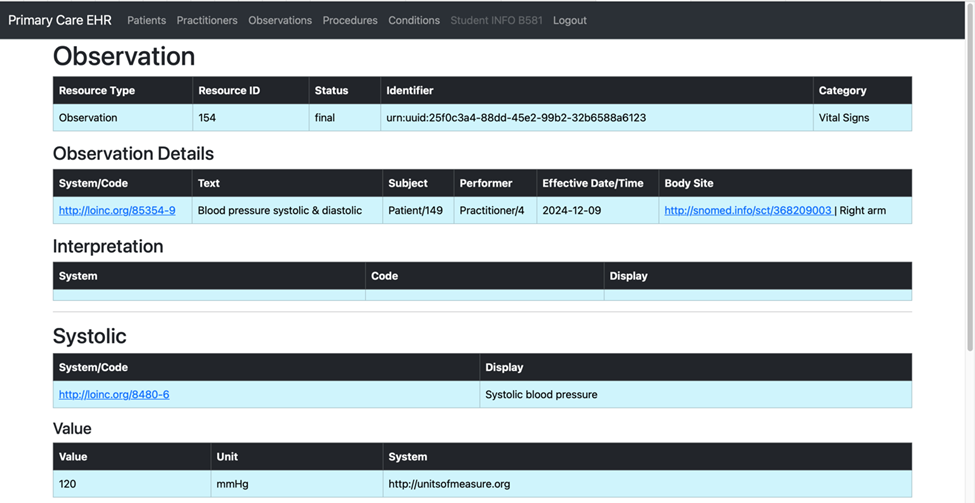
Task 3: Posting an Observation
In Task 3, we aimed to post an Observation for the previously created patient directly into the Primary Care EHR. Unlike the earlier tasks, this step did not involve querying the OpenEMR system. Instead, the observation data was manually prepared in a JSON file formatted to comply with FHIR standards.
The Python script utilized a function to read the observation data from the JSON file and another function to post this data to the Primary Care EHR's FHIR endpoint. This approach simplified the process by bypassing external queries and focusing solely on the transformation and loading of pre-formatted observation data, ensuring accuracy and seamless integration into the EHR system.
# Base URL of the FHIR server
BASE_SERVER_URL = "http://137.184.71.65:8080/fhir"
# Directory containing the JSON file
from pathlib import Path
import json
data_dir = Path.cwd() / 'data'
def read_data(file_name):
"""
Reads a JSON file
"""
json_file_path = data_dir / file_name
try:
with open(json_file_path, 'r') as file:
data = json.load(file)
return data
except FileNotFoundError:
print(f"File not found: {json_file_path}")
exit()
except json.JSONDecodeError as e:
print(f"Error decoding JSON: {e}")
exit()
def post_observation(file_name, resource_name):
"""
Posts an Observation resource to the FHIR server
"""
url = f"{BASE_SERVER_URL}/{resource_name}"
data = read_data(file_name)
{
"resourceType": "Observation",
"id": "149",
"meta": {
"versionId": "1",
"lastUpdated": "2024-12-06T12:00:00+00:00",
"source": "#1234XYZ",
"profile": [
"http://hl7.org/fhir/StructureDefinition/vitalsigns"
]
},
"text": {
"status": "generated",
"div": "<div xmlns='http://www.w3.org/1999/xhtml'>Blood Pressure Observation</div>"
},
"identifier": [
{
"system": "urn:ietf:rfc:3986",
"value": "urn:uuid:25f03c3a-88dd-45e2-99b2-32b6588a6123"
}
],
"status": "final",
"category": [
{
"coding": [
{
"system": "http://terminology.hl7.org/CodeSystem/observation-category",
"code": "vital-signs",
"display": "Vital Signs"
}
]
}
]
}
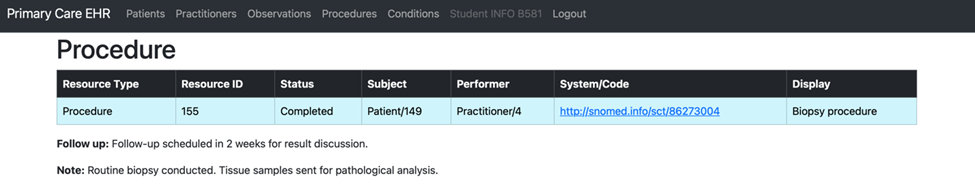
Task 4: Documenting a Medical Procedure
In Task 4, a medical procedure was documented for the created patient by crafting a JSON file in the required FHIR format. This file, procedure.json, was structured to include all necessary fields, such as the patient reference, practitioner details, and procedure code.
Using the TASK_4_post.py script, the JSON file was read and posted to the Primary Care EHR system, ensuring seamless integration of procedure details.
import requests
import json
from pathlib import Path
# Define the base URL of the FHIR server
BASE_SERVER_URL = "http://137.184.71.65:8080/fhir"
# Define the directory for storing data files
data_dir = Path.cwd() / 'data'
def read_data(name_of_the_file):
"""
Reads a JSON file and returns its content.
:param name_of_the_file: Name of the file to read (without .json extension)
:return: Parsed JSON content
"""
json_file_path = data_dir / f"{name_of_the_file}.json"
try:
with open(json_file_path, 'r') as file:
data = json.load(file)
return data
except FileNotFoundError:
print(f"File not found: {json_file_path}")
exit()
{
"resourceType": "Procedure",
"id": "149",
"status": "completed",
"subject": {
"reference": "Patient/149"
},
"performer": [
{
"actor": {
"reference": "Practitioner/4",
"display": "Dr. John Doe"
}
}
],
"code": {
"coding": [
{
"system": "http://snomed.info/sct",
"code": "86273004",
"display": "Biopsy procedure"
}
],
"text": "Biopsy procedure"
}
}
Challenges
Several challenges were encountered during the process of fetching the parent and child terms for conditions, primarily due to incorrect usage of endpoints. These issues were resolved by refining the ECL query and thoroughly testing the endpoints to ensure accuracy.
On the other hand, the authentication process and patient retrieval were executed smoothly, largely thanks to the exercises and guidance provided during class sessions. Ultimately, all tasks were successfully completed, and the loaded patient data, along with the associated conditions and procedures, were integrated seamlessly into the Primary Care EHR.